Processing a Cryo-Tilt Series with Fiducials
(IMOD 5.1)
University of Colorado,
Boulder
This is a guided introduction to generating a tomogram from a tilt series
of a cryo-sample with gold beads as fiducial markers for alignment. It
presents the most important concepts and details and provides brief explanations
of some points. It is based on an old and relatively low-resolution
example data set with a cellular specimen. For processing more modern tilt
series intended for subvolume averaging, you should go through
Processing a High-Resolution Cryo-Tilt Series with
Fiducials. For more details, consult the Tomography Guide, which you
can open from the Help menu in Etomo. It is also advisable to read through
relevant sections of the Tomography Guide before trying to process your own tilt
series. If you have not processed a tilt series in Etomo before, you may want
to do the dual-axis tutorial before this
example, especially if you have difficulty following the steps below without
shots of how the screen should appear.
The example data set is from a plunge-frozen PtK2 cell and was taken
with a Gatan Ultracam camera (a lens-coupled CCD camera with phosphor) on a Tecnai F30 by Cindi Scwartz.
First, a few points on conventions: labels in the Etomo or 3dmod interface are shown
in Bold, and entries in fields are shown in italics. For
mouse operations in the Zap window in 3dmod, the buttons are referred to as
"first", "second", and "third" because the buttons can be remapped in 3dmod.
If you have not changed the mapping, this corresponds to left, middle, and
right; otherwise, it refers to whatever you have chosen to be the first, second,
and third buttons.
Getting started:
-
Download the sample data set from our web site.
- Move the data set file "cryoExampleData.tar.bz" to the directory where you want to
work on it. Its contents will unpack into a subdirectory named "cryo".
- cd to the directory with the downloaded file
- Enter the command:
imoduntar cryoExampleData.tar.bz
or, anywhere except on Windows without Cygwin, you can use
tar -xjf cryoExampleData.tar.bz
- Enter the dataset directory with:
cd cryo
- Now you need to redefine the environment variable IMOD_CALIB_DIR to be the
current directory, which is simply "." (a period or dot), so that Etomo can find
some microscope-specific files used in the processing. Thus, you need to
enter a period at the end of all the command below to represent
the current directory.
- On Linux, Mac OS X,
or Windows with Cygwin, first give this command so that you will still have access
to parallel processing resources in case they are defined in a
system file:
cp $IMOD_CALIB_DIR/cpu.adoc .
Do not worry if there is an error; that just means you were not
using resources defined in such a file.
- On Linux, Mac OS X, or Windows with Cygwin, if you are using the bash shell or a related shell,
enter:
export IMOD_CALIB_DIR=.
Or, if you are use tcsh as the shell, enter:
setenv IMOD_CALIB_DIR .
- On Windows without Cygwin, enter:
set IMOD_CALIB_DIR=.
- Start the Etomo interface with:
etomo
Tomogram setup:
In this initial step, we define some features of the data set and create the
files needed for processing.
-
Press Build Tomogram in the Etomo "Front Page"
-
Press the yellow file chooser icon on the Datatset name line and select the stack file.
-
For SystemTemplate, select
cryoSample.adoc. This will set many parameters appropriately for cryo
reconstructions; you should generally select a template.
-
Press Scan Header to retrieve the pixel size and rotation angle of
the tilt axis from the image file.
-
Enter 15 for Fiducial diameter
-
Select Parallel Processing if you have more than one CPU core.
-
Select Graphics card processing if you have an Nvidia card with a GPU that can be used with IMOD.
-
For Image distortion field file,
select 27.5kGIF2007-03-24.idf. Data were taken with an older energy filter which introduced significant geometric distortions.
The distortions were measured and can be corrected with this file.
-
Press View Raw Stack and scroll
through the images to see that they are not well-aligned.
Notice the 4 contrast control sliders, which are present because the
data have been loaded as integers.
The top two sliders are very close together, which means the image data occupy a
very small fraction of the full data range; the rest of the data range is taken
up by image artifacts due to X-rays. In general, if you see bad views that you know
that you want to exclude from alignment and reconstruction, you can list
these in the Exclude views box.
-
Press Create Com Scripts
Pre-processing:
This step is needed to remove artifacts in the images, generally
produced by X-ray events in CCD cameras. These artifacts will produce streaks in a
reconstruction and can also make it harder to see the image features, which have
a much smaller dynamic range than the artifacts.
-
Press Show Min/Max for Raw Stack to
see the range of the data; both a plot of minimum and maximum values and a table
with more detailed information will open. Every view has a large negative minimum because there
was an X-ray artifact in the dark reference.
-
Press Create Fixed Stack to run the program that finds and erases
artifacts.
-
Press View Fixed Stack. Now there are only 2 contrast sliders because data
can be loaded as bytes after removing most of the artifacts.
-
Press Show Min/Max for Fixed Stack to
see the new range of data.
Deviations of 50-100 from the rest of the data will not matter in the
reconstruction.
- Press Use Fixed Stack.
- Press Done to advance to the next step.
Coarse Alignment
In this step, we use image cross-correlation to align successive images, which
makes it easier to track fiducial markers.
-
Press Calculate Cross-Correlation
-
When done, press Generate Coarse Aligned Stack
-
When done, press View Aligned Stack in
3dmod. Scroll through the images
to see that they now look aligned
-
Press Done to go on.
Fiducial Model Generation:
In this step, the positions of selected gold markers are found on all of the
images, which allows a more accurate alignment to be obtained.
-
Use Make seed and track and
Make seed model manually options. The option to make the seed model
automatically works quite well; we are not using it so that you learn how to do
this step manually if necessary.
-
Press Seed Fiducial Model. The Bead Fixer window opens in Make seed
mode with most of the settings that you need for adding seed points.
-
Turn on Automatic new contour in the Bead Fixer window if it is not on already. A contour is
a set of connected points. We need to put these points in separate
contours because a) we don't want to see them connected and b) each contour will
be added to with the corresponding points on other views.
-
Place the cursor very near each bead and add a point with the second mouse
button. It will be automatically centered (the Autocenter option is
turned on by Etomo in seeding mode).
-
Press the S key to save the model.
-
Switch to the Track Beads tab and press Track Seed Model.
-
The Project Log window shows the number of missing points when done. It
may be 0.
-
Press Fix Fiducial Model, which will load the tracked model into 3dmod and
switch the Bead Fixer to Fill gaps mode. You can scroll through the
views and also press the V key to see the tracks of the beads in 3D.
-
If there are missing points, you can now use the Bead Fixer to step from one gap
to the next (using the space bar as a hot key) and add a point, if appropriate. A bead does not need to be
marked on every view, and you should not add a point if the bead's position is
not clear. If you add points, be sure to save the model with the S
key.
-
Another way to complete the model is to press Track with Fiducial Model as
Seed. It is a good idea to look at the model in 3D first and make sure
there are not deviant points at the ends of contours that should be
fixed first.
-
Press Done to go on.
Fine Alignment:
Next the bead positions are fit to a mathematical model of specimen movements.
The model predicts a position for each bead on each view, and the mean distance
between the predicted and actual positions is referred to as the "mean residual
error". By obtaining a solution with some points left out, a
different error can be computed between the predicted and actual positions of
points not included in the fit, the "leave-out" error. It
requires many alignment runs with different points left out to a reliable
estimate of the leave-out error. Since this error is an estimate of how well the
solution generally predicts the positions of structures other than the beads
that happen to be included in the fit, it is a better measure of the quality of
a solution than the mean residual. The main problem with the mean residual
is that solving for more variables will always give a better fit to the points,
but using too many variables will fit excessively to the random errors in the
positions and do a worse job of predicting the positions of other points in the
specimen. You should always use the leave-out error to assess the value of any
change in the alignment parameters. However, either of these errors will let you
find and correct badly modeled points. .
The need to do so is reduced for some data sets by a method
called "robust fitting", which automatically gives less weight or even
eliminates the points most likely to be at incorrect positions. However,
it is good to learn how positions can be fixed manually.
-
Note that Do not sort fiducials into 2 surfaces for analysis was selected
by the "cryoSample.adoc" template.
-
Press Compute Alignment. When it is done, the Mean residual error
will appear in the Project Log.
-
Press View 3D Model to open the model of solved positions in 3D and
assess whether to solve for distortion (stretching and skew). Press the R
key in the Model View window for a side-view of the model. If you press the
middle mouse button and move the mouse to the right, and up and down a bit, you
can rotate the model until
the three lower beads are very close together.
-
To solve for distortion, fiducials should be well-distributed in Z: not all on
one plane, more than a few at a different Z height, and the ones at a different
height distributed over the area. This is not often the case for cryo data
sets, and this one fails the latter two tests.
-
Press View/Edit Fiducial Model. The Bead Fixer will open in (or
be switched to) Fix big
residuals
mode and read in the log from alignment. Zoom up in the Zap window to 2 or
so (more on a high-DPI monitor). It is also helpful to switch the Zap window to keeping the current
model point centered, by pressing the concentric squares icon in the toolbar.
-
If you press Go to Next Big Residual or the single quote (') key, it will
move to each point with a big residual in order by decreasing value. If a
point is not centered on the gold bead, you can move it to the correct position
by clicking with the third mouse button. The red arrow points to the
position predicted by the alignment solution; it is not based on any knowledge
of what is in the image. This position may not be
correct, but if it is, you can move the point to that position with
Move Point by Residual or the semicolon (;) key.
-
All of the biggest residuals occur at very high tilt, and each point has been
correctly modeled, so there is nothing to fix. The reason for this is
evident if you examine the output from the alignment program.
-
Right-click in Etomo over the Fine Alignment panel and select Align
log file. The error summary and the ratio of measured values to
variables solved for are in the Errors tab. Switch to the
Solution tab to see the alignment parameters.
-
Various columns from the solution table can be plotted by right-clicking in
Etomo and selecting a particular parameter. Right-click and select Plot
global mean residual. Notice that the mean residual gets much higher
at high tilt, which is why we saw only those points having the biggest
residuals.
-
To find possible erroneous points at lower tilts in this case, under Residual
Reporting, select Relative to Neighboring views.
Press Compute Alignment again.
-
There are no new points to fix.
-
Now that we have optimized the fiducial positions, we can optimize the alignment
parameters. First turn on Robust fitting with tuning factor
and press Compute Alignment.
-
The various error measures with and without weighting are summarized in the
project log. On the bottom line, the most important item about robust
fitting, the benefit, is enclosed in stars and indicates that it was
detrimental. Turn off Robust fitting.
-
Now press Run Cross-Validation. This runs the script
"restrictalign" that, as the name implies, starts with the current set of
variables and tests whether reducing the number or type of variables being
solved for will reduce the leave-out error and thus give a better solution.
-
When it is done, the results appear in the project log and any changes in
parameters are loaded back into Etomo. You should see that it changed from
solving for magnification and rotation at every view to solving for these
parameters in large groups, which means that there were only 2 or 3 values of
each being solved for, with linear changes being assumed within each group.
Such a result is typical when there are only around 10 fiducials.
-
Press Done to go on.
Tomogram Positioning
The goal of this step is to set angles and an offset in Z so that the specimen
is flat and centered in Z in the computed volume, thus minimizing the
computational effort.
-
Set Sample tomogram thickness to 500.
-
Press Create Whole Tomogram to build a whole, binned-down tomogram.
-
Press Create Boundary Model to open the tomogram
-
Open an XYZ window with Image-XYZ. Set Sum in the
toolbar to 20, which makes it average 20 slices.
-
The goal is to draw a pair of lines enclosing the sample at ~3 different Y
locations in the tomogram. For this specimen we can see the boundaries of the
sample well in cross-section, so this is the easiest way to draw them.
-
Use the Y slider in the toolbar to scroll through Y and get a sense of
how the boundaries change.
-
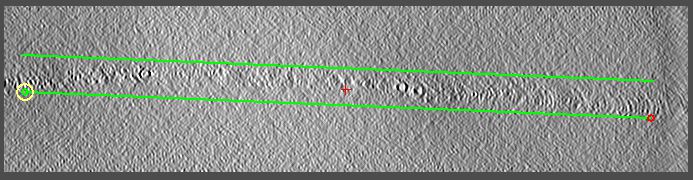
Go to the middle (~340) and draw two lines. 3dmod will automatically start
a new contour after two points. Move the current point with the third
mouse button; use the first mouse button to select a new current point. See the
image below.
-
Do the same near the lower end of the tomogram (axis position ~70) and the upper
end (~610). Your final model should 6 contours with 2 points each.
-
Save the model with the S key.
-
Press Compute Z Shift & Pitch Angles. The angle offset, Z shift, X-axis
tilt and tomogram thickness are shown; these will give the thinnest, most level
tomogram containing the area you outlined. The required thickness is also
shown.
-
Press Create Final Alignment, which reruns the fine
alignment.
-
Press Done to go on.
Final Aligned Stack Creation and CTF Correction:
-
Press Create Full Aligned Stack.
-
Switch to Correct CTF tab. Set Voltage (KV) to 300.
-
Press the file chooser icon on Config file and select F30_UltraCam_2008.cfg.
-
Set the Expected defocus to 8.0.
-
Press Run CTF Plotter. Ctfplotter loads the noise files and
then displays the power spectrum. The features of Ctfplotter are
explained in detail in the Guide
to Ctfplotter. In brief, the X axis is spatial frequency in
cycles per pixel, thus running from 0 to 0.5. The Y axis is the
logarithm of a rotationally averaged power spectrum, summed from
overlapping subareas referred to as tiles. The red or magenta curve
is the actual power spectrum; the green curve is a curve fit to it
based on equation that behaves like the CTF. The defocus value that
results from this fitting is shown in the top line after D:.
-
The low frequencies always dominate these power spectra, so the curve
has to be zoomed up to see the regions of interest properly. Draw a
rubber band to zoom up on part of the curve, such as frequency from
0.1 to 0.5, log power from -0.2 to 0.4.
-
The Fitting Params dialog lets you
adjust the range of points that will be fit. The Start fit at
value of 0.15 is reasonable; the End fit at value of 0.365 is
based on midway between the expected positions of the third and fourth zero;
this is also reasonable for the moment.
-
Set Baseline fitting order to 2 make the baseline flatter out to
0.4 and make it easier to fit to the peaks.
- Now that fitting is clearly functioning adequately, switch to Current defocus estimate in the
Angle Range dialog.
- Turn on Find astigmatism in the Fitting Params dialog.
The program fits to 90-degree wedges of data at all angles and finds the
astigmatism, which is substantial. You can set the Show wedge fits
value much lower, 0.001 to 0.005, and still see if the fits are good as they are
happening.
- With astigmatism accounted for, there appears to be useful data almost out to
the fifth zero. Double click with the right mouse button where the magenta
curve comes back to the baseline just before that zero; this sets the
End fit at value to ~0.42.
- Turn on Do auto weighting and truncation in the Fitting
Params dialog. This option makes it reduce the fitting range or give less
weight to weaker parts of the curve, based the correlation between green and
magenta curves, so you do not have to adjust fitting range for weaker signals.
The light blue curve shows the weighting value.
- Set Number of Views to fit to 1. The spectrum is much noisier but the
fit to the fifth zero seems reliable. Given this amount of noise even at
zero tilt, it is a good idea to step through some of the tilts to see if the
fits will still be good at higher tilt.
- Enter 5 in the Step view range by box. Press
Step Up repeatedly. The fit deteriorates suddenly somewhere around 25
to 30 degrees and then recovers above that, probably due to the directionality
of useful signal in this specimen. (The angle where this happens depends on what
angle offset you ended up with in positioning). Increasing the
Number of views to fit to 3 restores the quality of the signal enough
to allow good fitting. Set it back to 1 after observing this.
- Somewhere around the same range of angles, the fits to some of the wedge
spectra look bad, and the astigmatism estimate jumps to a high value (1.6-1.8).
This also happens at the highest negative tilt. The best remedy for this
is to set Min views for astigmatism to 11.
- Return to zero tilt by setting Middle tilt angle to 0.
- In the Autofit section, check the box to Fit 3 views
above and set the threshold angle to 25. This option has
an effect only when autofitting. Check Fit every angle: step by 1.
- Press Autofit All Steps. The wedge spectrum fitting
should look well-behaved at all tilts.
- Press Graph Values to see curves for defocus and the
astigmatism amount and angle versus tilt angle. Astigmatism peaks at
around 35 degrees. Find the view in the table with highest astigmatism
value and double-click that line to load the spectrum there. Press Apply
to recompute the spectrum and confirm that the fitting to the wedge spectra does
not jump to aberrant values.
- You could do the fitting manually to use more views for astigmatism and
defocus only where needed, but this is all the effort these low-resolution data
are worth. Press Save to File and exit Ctfplotter.
- Select up to 12 CPUs
or up to 4 GPUs in the parallel processing table.
- Press Correct CTF then Use CTF Correction when it is
done.
Gold Erasing:
Gold beads are by far the densest items in cryo-reconstructions and they
cast artifactual rays that are about as dense as the biological features.
To minimize this effect, it is often desirable to remove the beads from the
projection images before reconstruction.
- To erase gold, select the Erase Gold tab.
- Select Use findbeads3d to find the locations of the gold in a
binned-down tomogram and project their positions onto the tilt series images.
-
Set the Thickness to 220.
-
Press Align and Build Tomogram.
-
When it is done, select Store only points above threshold and press
Run Findbeads3d. This will find the beads in the tomogram as well as
it can, and only put the points into the model that have "peak strengths" above
what it thinks is the best threshold between beads and non-beads.
-
When it is done, press View 3D Model on Tomogram. The Bead Fixer
opens with some special controls for adjusting the threshold peak strength, in
case we had stored more points in the model.
-
Press the V key to open the Model View. If you see only 10 of the 11 model
points (because of a bug in Findbeads3d) push the Threshold slider to the
left. Put the mouse near one of them and click the right button; the
Zap window will change to show that model point on the image. If
necessary, you could add or delete points in this model. Close 3dmod.
-
Press Reproject Model then View 2D Model on Aligned Stack.
Look at the high tilt views; you will see that some of the points are not
well-centered.
-
Change two parameters to compensate for this poor centering: set Diameter to
erase to 26, and turn on Iterations to grow circular areas and
set the value to 3.
-
Press Erase Beads. When it is done, press View Erased Stack
to see the result.
-
Press Use Erased Stack.
-
Press Done to go on.
Tomogram generation:
At last, you can compute the tomogram.
-
Make sure only one of the two choices Use the GPU and Parallel
processing is selected if you only have one GPU, and press Generate Tomogram.
-
Press View Tomogram in 3dmod when it is done.
-
Press Done to go on.
Post-processing:
In this step, you can trim away unneeded regions, convert the tomogram to bytes
to save time and space, and reorient the tomogram so that the slices stored in
the file are in X/Y planes instead of X/Z planes. Even if you do not
want to trim or convert to bytes, you should always go through this step to get a
reoriented tomogram, which will work better with other
programs.
-
Turn on the rubber band in the Zap window and select an area of about 1500x1500
pixels, i.e., at a window zoom of 0.5, the rubber band size on the screen of
about 750x750 pixels, as shown in the Zap toolbar.
-
You can move an edge or corner of the rubber band with the first mouse button,
or shift the whole band by pressing the second mouse button with the cursor over
an edge.
-
Scroll to the first slice that you want to keep and press Lo, then find the last
slice you want to keep and press Hi. To make a file suitable for
practicing NAD filtering, keep 60 slices or less.
-
In Etomo, press Get XYZ Volume Range from 3dmod to import these limits.
-
In the Zap window, select a narrower range of slices with Lo and Hi
that will be used to determine scaling; exclude the dense contamination on the
surface.
-
In Etomo, press Get XYZ Sub-Area from 3dmod in the Scaling section
to import these limits.
-
Press Trim Volume. When done, press 3dmod Trimmed Volume to
see the result.
-
Press Done to go on.
Automatic Seed Model Generation:
Making the seed model manually was just an training exercise; you should be able
to make this model automatically with almost all of your own data sets.
For an example of doing this, return to the Fiducial Model Generation
page.
-
You may first wish to save the files from your manual work (cryo.seed and
cryo.fid) by copying them to a different name.
-
Switch to the Seed Model tab and select Generate Seed Model
automatically.
-
The one required entry is either a number or density of beads to select.
Enter 25 for Total number. You can enter a standard number
appropriate for the size of the images, even there are not nearly that many
beads. It will find as many as it can, but not select more than the
indicated number.
-
The option to Use boundary model is available in cases where you do not
want fiducials outside of a defined area. In cryoET, it is usually
desirable to avoid using beads over the carbon, if possible.
-
Press Generate Seed Model.
-
When it is done, the project log shows the essential results: there were 10
candidate beads, 1 was considered too elongated to use, and it thus picked 9.
-
Press Open Seed Model to see the points picked. You will have to
page up one view to see them. One bead was eliminated from consideration
because it was too near the left edge. The other bead skipped is in the
upper left quadrant. It looks fine at zero degrees but is actually two
beads on top of each other and tracks poorly at higher tilts. The program
eliminated it because it detected sufficient elongation in the lower tilts that
it examined.
Clean Up:
In most cases, there is no need for the intermediate files from processing.
This step allows you to remove these files and leave all of the information from
which they could easily be recreated if necessary. The original raw tilt
series stack can also "archived" by compressing its difference from the current
stack; this operation is reversible.
- Press Archive Original Stack and confirm the deletion of the original
stack when it is done.
- All intermediate files are now shown in the box. Click in the box and
type Ctrl-A to select all files. You could then click on individual files
while holding down the Ctrl key to unselect them. You might wish to leave
cryo.preali (coarse aligned stack), cryo.ali (final aligned stack), or
cryo_full.rec (raw reconstruction, which could be trimmed differently).
- Press Delete Selected to remove the intermediate files that are still
selected.

