Click image for full-sized view.
Download and Installation Page
Tilt Series Acquisition with SerialEM
Single Particle and Multiple Grid Acquisition with SerialEM
Program Features
Contribute to the Continuation of SerialEM
Highlights of SerialEM 4.2
Availability of SerialEM for Various Hardware
Configurations
Setting Up SerialEM
How to Report a Problem with SerialEM
SerialEM Discussion Group
Other Helpful Materials
Source Code for SerialEM, SerialEMCCD Plugin, and Python Module
Mirror Site for SerialEM
SerialEM is a program that can acquire a variety of data from electron microscopes: tilt series for electron tomography, large image areas for 3-D reconstruction from serial sections, and images for reconstruction of macromolecules by single-particle methods.
The main focus of the program is automated tilt series acquisition. For this, it uses an approach based on prediction of specimen position during the tilt series from the position at previous tilts. It does not count on the microscope or the specimen being particularly well-behaved, so unless a prediction appears reliable, it falls back to measuring and adjusting defocus and/or specimen position when necessary. With this method, it achieves both robustness and speed. For example, a 2Kx2K, 1 degree tilt series can be acquired in about 20-25 minutes with a four-port readout CCD camera.
The method and the program are described in this
paper:
Mastronarde, D.N. 2005. Automated electron microscope tomography using robust
prediction of specimen movements. J. Struct. Biol. 152:36-51.
The prediction method was first described in this
abstract from the 2003 MSA meeting,
Mastronarde, D.N. 2003.
SerialEM: A program for automated tilt series acquisition on Tecnai
microscopes using prediction of specimen position. Microscopy and
Microanalysis Vol. 9, Suppl. 2, 1182CD.
SerialEM is also widely used to acquire data for single particle reconstruction. After positioning on a hole, it can acquire multiple images in a hole and/or in multiple holes using image shift, with compensation for the beam tilt and astigmatism induced by the image shift. There is a routine that finds hole positions automatically and one that can combine them into an optimal subset of positions for acquiring from multiple holes. The basic framework for this automated acquisition is through the Navigator module, which has interface for acquiring from the selected points and performing all commonly associated actions.
SerialEM 4.2 added the ability to acquire automatically from a series of grids on a microscope with an autoloader. The interface for multiple grid operations gives flexible control over the sequence of steps from initial mapping of the whole grid to final acquisition.
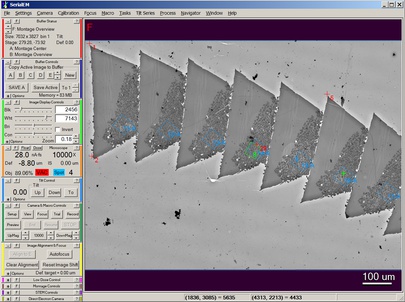
In addition to automating data acquisition, the program provides an integrated user interface for image acquisition, display, and storage. It looks like this:

Other notable features include:
SerialEM was originally developed with grant funding from NIH. Since 2014, it has been maintained and developed with funding that comes from users, which has allowed us to focus on coding and documenting instead of grant-writing. Since 2018, funds have also been used to pay Guenter Resch to provide user support.
You can help make SerialEM a community-supported resource by having your laboratory or microscope facility contribute to the further development and continued availability of the program. The mechanism for doing this is to purchase priority annual support, so that the expense represents a tangible item that can be paid from grant or institutional funds. In September 2020, we transitioned to having these payments made to Guenter Resch's company, Nexperion. In addition to directly funding Guenter's assistance to users, The majority of the funds are used to compensate David Mastronarde for his work on SerialEM, with the rest directly funding assistance to users by Guenter and by Chen Xu (at the University of Massachusetts Chan Medical School). We offer three options, allowing you to pick the size of your payment based on your financial resources and the importance of SerialEM to your work. The options are:
What happens if you do not pay? Although the first option represents the kind of support that we provide routinely, payment for this support is voluntary. We do answer questions and deal with problems from non-paying users, but may take longer to do so.
Why is it important to contribute? User contributions are important both financially and because they are not tied to specific developments and represent the general user interest.
What does the change in 2020 mean for the future of SerialEM? This is not a move toward commercialization. We intend to keep the program freely available. It is already open source.
How do I get help with SerialEM under the new arrangement? For everyone, requests for support and for initial configuration (framework) files should still be sent to serialemsupport@colorado.edu. (See How to Report a Problem with SerialEM).
Click Here to go to the registration page for support agreements on Nexperion's web site. (This initial registration is required only once.)
The most notable features of the latest release, SerialEM 4.2, are:
There are many other improvements; see the list of notable features to learn about them.
SerialEM works on modern Thermo Fisher (FEI) microscopes (Tecnai, Titan, Spirit, Talos, Glacios), JEOL microscopes with the TEMCON or TemCenter interface (e.g., 1400, 2100, 2200, 2500, 2800, 3100, 3200, ARM200, F200, CryoARM), and the Hitachi HT7700 and HT7800. The program is built and tested primarily with a Tecnai simulator on Windows 10, but it still works on Tecnais under Windows 2000.
SerialEM works with CCD or CMOS cameras from Gatan, TVIPS (Tietz Video and Image Processing Systems), Thermo Fisher, AMT, DirectElectron, and EMSIS (formerly OSIS, Olympus Soft Imaging Solutions); it also supports the JEOL Ruby and other cameras. Direct detection cameras from Gatan, Thermo Fisher, and DirectElectron are supported. The EIGER2-based EM detectors from DECTRIS are also supported through a plugin currently available from DECTRIS. All EMSIS cameras work through a plugin available from EMSIS for either 32- or 64-bit versions. STEM acquisition is supported from Gatan's DigiScan, Thermo Fisher's STEM imaging, the TVIPS scan generator, and the JEOL STEM on the 1400 series, F200, CryoARM, and possibly other systems with similar interfaces.
If necessary or advantageous, SerialEM can be run on a separate computer and control the microscope over the network. Such a configuration is standard on JEOL microscopes, is quite common when using DE and newer Gatan and Tietz cameras on Thermo Fisher microscopes, and is also possible for Hitachi microscopes. The program can access Gatan, Direct Electron, Thermo Fisher, and Tietz, and JEOL cameras over the network from another computer, but for other kinds of cameras, SerialEM must run on the computer that hosts the camera.
For running on a Thermo Fisher microscope, it is necessary to have the Thermo Fisher scripting adapter installed. If you do not have the adapter, you need to purchase it from Thermo Fisher. To check whether it is installed, do the following:
Although SerialEM itself is open source, the microscope plugins and some camera plugins in this software are not. The University of Colorado retains various rights with regard to these components; however, they are currently provided free for both commercial and non-commercial purposes. By downloading the distributed software, you agree that you will not distribute it outside of your own institution.
The program has been installed on over 400 Thermo Fisher and 200 JEOL microscopes.
Program and configuration files are available on the Download and Installation page. Follow the instructions in there to get started. As a first step, fill out the questionnaire in Word doc form and send it to serialemsupport@colorado.edu; we will prepare an initial configuration file. This is strongly recommended for TFS/FEI scopes and absolutely essential for JEOL and Hitachi scopes. Installations on new JEOL microscopes are usually done with the assistance of Jaap Brink from JEOL (jbrink at jeol dot com), but sending in the questionnaire is still the starting point.
Guenter Resch from Nexperion offers
SerialEM installation and training for both Thermo Fisher and JEOL microscopes. This
service is available worldwide, on site or remotely.
Chen Xu at the University of Massachusetts Chan Medical School and Gabriel Frank at
Ben Gurion University in Israel may also
be willing to help with installation; email

for contact information.
Send all problem reports and requests to serialemsupport@colorado.edu. This is a private email address, not a mailing list. Guenter Resch is the primary contact for getting assistance with SerialEM. He will collect needed information about a problem, and handle most problems with use of the program or pass them on to Chen Xu. Problems in the program itself will be handled by David Mastronarde.
What we can support: We can only diagnose problems that occur in relatively recent releases. If you are running a version prior to the current stable release version (e.g., 3.8.10, or a 4.0 beta version, after 4.0 is in stable release and 4.0.x packages are available), you must upgrade and see if the problem still occurs. If you are running the current stable release version but it is not the latest update (e.g., you have 4.0.5 and the latest is 4.0.12), you should upgrade before reporting the problem. If you are running a beta version in the current development cycle that is not based on the latest beta release package, upgrade if possible, or at least check the History-beta.txt or History-testing.txt file before reporting the problem.
The following information and files can help to address your question faster and more effectively. Here are the essentials:
These additional points can also be helpful:
Please follow these additional tips for effective communication:
The SerialEM discussion group provides a means for users and people managing the program to ask questions and share their experiences, as well as a way to receive notification of new releases and other news. Digests and posting via email (i.e. list server functionality) are provided. Additional capabilities, including browsable / searchable archives are available through a web interface. Access to the web interface requires a Google login profile, which must be associated with the email address at which you wish to be contacted. (Non Gmail addresses can be used with a Google login, with or without a Gmail address. See the Email section of your Google Account settings). The postings are not moderated, but joining requires approval.
To subscribe or visit the group via the web interface please visit
SerialEM.
If you do not have and are not willing to create a Google
login and are satisfied with email only access, you can also request
membership by sending an email to

asking to be added to the SerialEM list, and you will be added
manually. Be sure that this email is sent from the same address as you
would use to post to the list.
After your request is approved, you will be added to the group and will receive a confirmation email with additional instructions. Once subscribed, we request that you edit your membership settings for this group so that your display name is your real name. To post to the forum you can either use the web interface or simply email your posting to serialem@colorado.edu
To cancel your subscription, use the web interface or send an email to serialem+unsubscribe@colorado.edu.
When replying to a previous posting via email, you will have the option to reply to the sender or to the list.
A standalone copy of 3dmod and associated files that can be run on Windows without installing IMOD or Cygwin is available here. The package unpacks to C:\Program Files\3dmod by default and a shortcut can be set up to run 3dmod from there. The package also includes midas, which can be useful to have on the microscope for checking rotations between magnifications. The latest package includes ctfplotter for determining defocus and astigmatism, and genhstplt for displaying graphs, and SerialEM 4.1 onward includes commands to use these.
Fixed-width font narrower than Courier. Place in C:\ProgramData\SerialEM or C:\Program Files\SerialEM
An optional plugin to SerialEM can provide direct access to JEOL microscope functions through the TemExt interface without relying on Python. It enhances SerialEM scripting capabilities and allows for an event-driven scripting approach. It is available here. See the README.txt file for requirements.
A repository for contributed scripts was developed by Guenter Resch from Nexperion and is hosted on the Nexperion site. If you have scripts that may be useful to others, please consider adding them to the repository.
Guenter has also developed a web page on the Nexperion site that can prepare a Basic mode file for a particular set of activities, so that controls not relevant to those activities can be hidden.
Daniel Caujolle-Bert at the University of Basel has written a program for managing user and settings and scripts in an environment where multiple users share the same login; it is available here.
Guenter is creating a new set of video tutorials on particular operations; so far they include autocontouring grid squares and backing up a SerialEM installation. These are available on our Vimeo page.
Our YouTube channel has a series of videos of the lectures on SerialEM presented in a webinar in September 2012, plus some tutorial videos prepared by Cindi Schwartz. The videos are also available right here; if you sort on the "Last modified" column they will be in a good order for viewing.
Here are some training documents written by Cindi Schwartz, to provide concrete guidance for doing room temperature and cryo tomography.
Here are Powerpoint presentations from a workshop on SerialEM given in December 2006. The slides have a lot of words, but there are many fewer words than in the help manual, so they may be helpful by highlighting the most important points.
And again, here are the:
Both SerialEM and the SerialEMCCD plugin to DigitalMicrograph are open source;
SerialEM under an MIT license, and SerialEMCCD under the GPL version 2. You can
browse the SerialEM source here or
on Github and
SerialEMCCD source here or
on Github, or you can download these
repositories with Mercurial or git, repectively. The primary repositories for
the SerialEM components available here are now git
repositories on our private server; the repositories linked here are
all updated nightly and when a Testing executable is released.
See the IMOD source
instructions for setting up Mercurial and updating a repository.
To clone the SerialEM repository with git, run
git clone https://github.com/mastcu/SerialEM
To clone it with Mercurial, run
hg clone https://bio3d.colorado.edu/SerialEM/SerialEM
Older versions of hg may require "hg clone --insecure" here and in the other clone commands below.
To clone the SerialEMCCD source with git, run
git clone https://github.com/mastcu/SerialEMCCD
To clone it with Mercurial, run
hg clone https://bio3d.colorado.edu/SerialEM/SerialEMCCD
If you are going to try to compile either of these projects, you will also
need the collection of libraries, which is in a separate
SerialEMLibs repository here or
on Github. You can
download this with git with:
git clone https://github.com/mastcu/SerialEMCCD
To clone it with Mercurial, run
hg clone https://bio3d.colorado.edu/SerialEM/SerialEMLibs
The module for Python support is also available here. You can browse
the project files here
or on Github or you can clone the
repository with git:
git clone https://github.com/mastcu/PythonModule
To clone it with Mercurial, run
hg clone https://bio3d.colorado.edu/SerialEM/PythonModule
Occasionally, this site becomes inaccessible for various reasons. There is a mirror site available at https://bio3d-mirror.nexperion.net