The interface is organized into two tabs. The Input and Pre-processing tab has entries to specify the frame stacks to process, various ways to enter tilt angles and other metadata, and gain normalization, defect removal, and truncation of the input values. The Alignment tab has entries to control the alignment process and output files.
Input file specification
This section provides three different ways
to specify what frame files to process.
After selecting file(s) in this section, the Directory field will be set with the working directory where Alignframes will be run and output files will be created. This will be the directory of the ".mdoc" or file list file, or the common directory where you selected frame files. If you have an ".mdoc" file and the frame files are not in this directory, then you need to switch to Advanced mode and select the directory with frames in the Other directory with frames field.
The Root name for output files will be the basis for the names of all files created in processing. It will be initialized using the name(s) of the file(s) selected in the input section, but you are free to change it.
Other sources of metadata
The set of options in this section are
enabled when input files are not being taken from an ".mdoc" file. Their
primary purpose is to get tilt angles and the tilt axis rotation angle into the
output tilt series stack. However, they can also be important for enabling
dose weighting with a fixed dose per tilt image.
There are two rules that must be followed when using the options in this section (the programs cannot check whether your data comply). First, if the tilt angles are not being extracted from filenames, the order of input files needs to match the order of provided tilt angles. Second, if dose weighting is to be done with a fixed dose per image, the input frame files must be in the order in which they were acquired. Note that dose weighting is enabled when either Text file with tilt angles or Tilt angles in filenames is selected, but not when a tilt series is the sole source of angles, because the order of a tilt series is unlikely to match the order of acquisition.
Reading EER Files
Your choice of how EER files are read determines the size and number of
frames that Alignframes deals with. The default is to sum frames to produce
12 images to align for each tilt angle, and to read frames with
reduction to 8K, where this summing and reduction is done efficiently in the
TIFF file reading module. The resulting pixels are the "unbinned pixels"
referred to elsewhere and used as units for specifying filters and reporting residuals.
Note that as long as you are
gain-normalizing, the reduction to 8K or 4K frames is done with antialiasing,
so it will not introduce significant aliased noise. If you ultimately want 4K
output, the best strategy is to use the default reduction by 2 on input, as
the second reduction by 2 will be done by Alignframes with Fourier cropping
for even better antialiasing. Nevertheless, the penalty if you need to
read in frames as 4K is not as substantial as it would be if ordinary binning
were used.
If want to change these defaults, open the section Reading EER files4K or as 16K. You can change the number of images to align in the Sum frames to make spin box, or you can switch to Sum successive sets of frames and directly specify the number of frames to sum when reading.
Gain normalization
If you have unnormalized frames from a K2 or K3 camera, or EER files from a
Falcon 4 camera, the option Gain
normalize from reference and defect files in frame file header provides
the most convenient way to handle normalization. For a K2 or K3 frames,
Alignframes will use the
gain reference file listed in a title in the header, and a defect file if any, and also
apply whatever rotation and flip to the reference is indicated by the "r/f"
value in the title line. For an EER file, the program will find the
name of the gain reference in the metadata under tag 65001. The reference
and defect files must be in the
directory with the frame files. If you need something besides this behavior,
switch to Advanced mode, where you can make separate entries for the
two files and for the rotation and flip operation. If you have the option to
use values from the file header checked, any of these advanced entries will
override the respective item from the header.
Truncation
You can choose to truncate high values, which can help prevent hot pixels with
many counts from affecting the alignment. The first option is to truncate at a
given number of counts, which is useful when you have raw electron values but
more difficult to work with when you have scaled, normalized values. With raw
values, you can use the command "clip hist" on a frame file to see where the
main distribution falls off and the tail of hot counts begins. The
second option lets you specify the number of standard deviations above the mean
at which to truncate, which will work with all kinds of data. Here a value of
10 or more may be appropriate.
This tab of the dialog has options to control the frame alignment itself, dose weighting, and output size, scaling, and orientation. Some of the defaults in this section are set based on the assumption that computational time is of minor importance, unlike the situation when aligning frames during acquisition.
Fitting to pairwise shifts
The basic method of alignment is to use cross-correlation to find the shift
between many pairs of frames, not just successive frames. Linear equations
are fit to these pairwise shifts to obtain the shifts that must be applied to each
individual frame. Because of the multiple measurements, robust fitting can be
used and pairwise shifts that appear to be statistical outliers are
down-weighted or ignored entirely. The default option is to do one robust fit
to the pairwise shifts between all pairs of frames. For example, if there are
14 frames, then there are a total of 91 pairwise shifts measured, and the
linear fit
solves for 14 frame shifts whose sum is constrained to be 0. Thus there are only 13
independent variables, and the ratio of measurements to unknowns is 7.
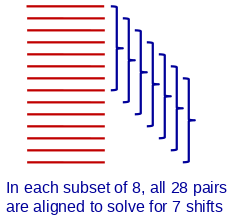
The other two options in this section fit to pairs from subsets of the frames, a strategy that keeps alignment time from depending on the square of the number of frames. The diagram illustrates this situation with 14 frames, and sets of 8 being fit at a time.

The option to fit to half the frames can be useful when aligning higher-dose images for single-particle reconstruction, where it avoids correlating frames that may be too dissimilar because of changes in the specimen during the exposure. For 40 frames, 570 pairwise shifts would be measured.
The default, fitting to all the frames, is both the most time-consuming and the one with the highest ratio of measurements to unknowns. It thus has the best chance of giving an accurate result with difficult alignments having a low signal-to-noise ratio. For this reason, the default in the iterface is to fit to all the frames.
Reduction and filtering
The initial image reduction and low-pass filtering in Fourier space work
together to filter out noise so that the true correlation
peak can detected. The reduction is referred to as binning, but is in fact
antialiased reduction to avoid introducing more noise. The reduction is in a
sense dispensable, because the same accentuation of the correlation peak could
be achieved with a filter alone, but some reduction is
essential to keep both the memory usage and the time for the many pairwise
correlations reasonable.
The Basic-mode choices for reduction are either Default, Reduce by N (a specified binning value), or Reduce to about N pixels, in which the integer binning will be used that make the size come out closest to the specified target. The Default choice will reduce to a somewhat flexible preset target size, either 1250 pixels, or 1560 pixels for frames recognized as coming from a K3 camera. The log from Alignframes will report what binning was selected when reducing to a target size.
A filter cutoff is a frequency at which a Gaussian rolloff starts to be applied to image. Frequencies are specified in unbinned reciprocal pixels so that the filter effects will be relatively invariant with different binnings. In these units, the highest frequency in the unbinned image is 0.5/pixel along an axis or 0.71/pixel along a diagonal. After a reduction by factor N, the highest frequencies retained in an image being filtered are 0.5/N /unbinned pixel along an axis and 0.71/N along the 45-degree diagonal. With a binning of 6, these correspond to 0.083 and 0.118/unbinned pixel, which means that a filter with a cutoff of 0.1/unbinned pixel will have very little effect.
Multiple filters can be applied with very little additional computational time, and the shifts from whatever filter gives the best overall fit will be used. Here, the best fit is assessed from the "leave-out error" rather than the mean residual, which reflects how well a solution obtained by leaving out one or a few of the pairwise shifts can predict the shifts left out. The default is to use multiple filters spanning a broad range; the comma-separated list is placed in the Filter cutoffs text box. Using multiple filters is particularly appropriate for processing tilt series because frames at higher tilt may have less contrast and need stronger filtering. With multiple filters, there will two lines at the end of the Alignframes log showing the number of frame stacks that came out best with each filter.
The Use hybrid shifts option is relevant only when there are multiple cutoffs and fits are being done to subsets of frames. For each subset of frames, the program uses the shifts from whatever filter gives the best fit on that subset. The reasoning is that this method selects shifts from the fits with the lowest errors.
Grouping to Improve SNR
Sometimes the frames are just too noisy to correlate reliably with each
other. Grouping frames by selecting the Group frames checkbox can
overcome this problem by increasing the SNR of the images being correlated.
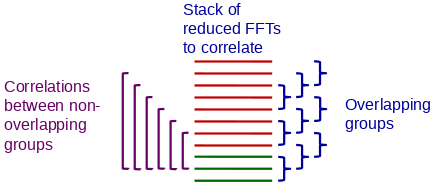
Grouping would allow you to take advantage of the increased temporal resolution of EER files by specifying reading in with more frames to align. Since overlapping groups are analyzed, a shift can still be found for each frame. For example, if you have 36 frames, grouping by 3 would make the SNR when doing pairwise correlations be the same as if the data were read in as 12 frames. However, you may also need to select Refine in groups for the refinement step.
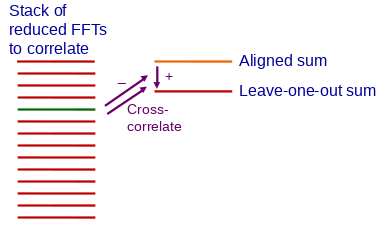
Refining the Alignment
Alignframes can refine the alignment produced from analysis of pairwise shifts
by correlating each frame with the sum of the rest of the aligned frames.
Spline smoothing
After the shifts are calculated, they can be smoothed by fitting a spline curve.
This smoothing is set to occur by default when there are at least
20 frames. It may be useful down to about 15 frames, but it is advisable to
evaluate the output in some way before using it for fewer than 20 frames. One
way to compare results with and without smoothing is to look at the FRC
(Fourier ring correlation) output in the log when Alignframes is run with the
"LinesOfAlignSummary 3" option.
Dose weight filtering
Dose weighting can be done using either doses from an ".mdoc" file or by
entering a fixed
dose per summed image. An ".mdoc" file is ideal because it contains information
that allows
the accumulated dose to be known for each image in a tilt series.
In order for dose weighting to work properly without an ".mdoc" file,
two rules must be followed:
Select Do dose weighting and the section will open up. Select whether to use the doses from an ".mdoc" or a fixed dose, and in that case enter the fixed dose in the text box. Even when there is an ".mdoc" file, you can enter a fixed dose to supercede the dose values in the file; the dose you enter, as well as the new values for the accumulated dose at each frame, will be inserted into the adjusted ".mdoc" file written by Alignframes.
With the option Normalize within each set of frames selected, the filters within each set of frames are normalized to add up to 1 at all frequencies. High frequencies will be attenuated for later frames and actually boosted for the earlier frames, and there will be no overall attenuation through the tilt series. Normalizing is on by default to fit the recommended workflow within Etomo, in which the real dose weighting between tilt images is done on the final aligned stack. The output with normalization will perform better in bead tracking and CTF detection. (If bead tracking benefits from filtering, use the uniform filtering provided there rather than the dose-dependent filtering of dose-weighting.) The normalized weighting is in fact a minor modification of the data that will be significant only for the early tilt images. If you turn off this option, you can select Non-dose weighted output also in Advanced mode to get an unweighted stack also.
Check Microscope voltage is 200 kV for images taken at 200 kV.
Other Basic mode options
To make aligned sums reduced in size with Fourier cropping, select the desired
reduction with the Reduce output size by spinner.
Check Use the GPU to use an NVIDIA GPU for processing. The processing itself may be about 5 times faster, or more, but with image input and output taken into account, the overall speedup may be only a factor of 2-3.
Action Buttons and Results
Press Run Alignframes to run the process.
Press Plot All Results to open all three of the graphs available, which can also be opened individually from the right-click menu. The graphs are:
Press Open Output Tilt Series to open the stack of aligned sums.
Press Start Reconstruction to open the Setup Tomogram interface for this stack in another tab in Etomo. A directory chooser will appear first to allow you to select a directory in which to do the reconstruction. You can create a new directory there if needed. Both the aligned stack and a new, adjusted ".mdoc" file, if any, will be moved to the directory.
The output tilt series is named "rootname_af.mrc" if no dose weighting is done or it is done with normalized filters, or "rootname_af_dw.mrc" if it is a dose-weighted without normalizing. If an ".mdoc" file was provided, the adjusted file will be named the same as the output stack, with ".mdoc" appended. If Non-dose weighted output also was selected, then the unfiltered stack is named "rootname_af_nodw.mrc"